AUTOCROP
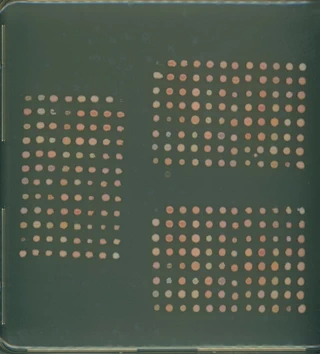
In high-throughput microbial research, monitoring growth rates across hundreds of samples is a significant logistical challenge. This project was developed to automate the processing of images for Rhodotorula colony growth assays.
To maximize efficiency during data collection, multiple 96-well plates (typically three or four) are scanned into a single large image. However, to analyze growth rates, each plate must be isolated and standardized so that images of the same plate can be compared across different time points. AUTOCROP is a Python-based solution that replaces manual cropping with an automated pipeline.
The Challenge
Standardizing colony images involves overcoming several environmental variables:
Color & Exposure Standardization: Because scanners automatically adjust color and contrast based on the density of the sample, images taken at different time points often lack consistency.
Dynamic Growth Sensitivity: Early-stage or low-density colonies are often too faint for standard edge-detection algorithms to identify.
Key Features
Here is the refined and formatted Features section for your project documentation:
Color Normalization & Pixel Consistency: To enable precise growth rate analysis, the pipeline includes a standardization step that corrects for automatic scanner exposure and color shifts. This ensures that pixel-to-pixel comparisons remain accurate across images taken at different time points throughout the experiment.
Dynamic Parameter Sets: To handle the “invisible colony” problem, the tool includes three sensitivity presets (High, Medium, and Low) tailored to colony density. These adjust the adaptive thresholding logic so that even low-biomass, faint plates are captured without losing their geometric boundaries.
Auto-Detection & Manual Overrides: By default, the script analyzes the image context to automatically select the optimal processing parameters. If unique lighting artifacts or experimental glitches occur, users can easily force a specific preset via command-line flags (by using different preset).
Flexible Layout Configurations: While optimized for the standard 3-plate scanning template used in our Rhodotorula research, the tool supports custom plate counts.
Research-Ready Batch Workflow: AUTOCROP is designed to be wrapped in simple bash loops, allowing for the fully automated processing of entire data directories. This transforms a bottleneck of thousands of raw scans into analysis-ready files in minutes rather than hours.
If you’re interested in AUTOCROP, please refer to the GitHub page for more details.
