Metagenomics
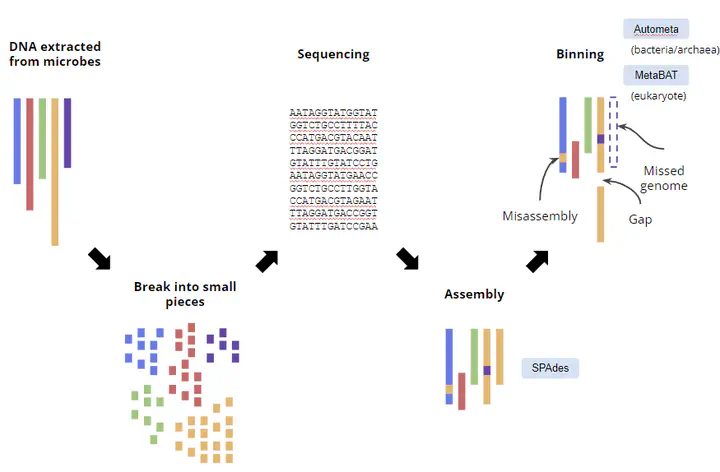
There are 2 major metagenomic approaches to investigate the microbiome - amplicon sequencing and shotgun sequencing. The major aim for amplicon-based approach is estimate the relative abundance of each species in the sample. For shotgun-based method, we can retrieve the metagenome-assembled genomes (MAGs) and further interrogate the functional capacity of them.
Currently, we apply metagenomic techniques to 2 ongoing projects - The citrus huanglongbing (HLB) project and elk coprophilous microbiome project.
In the citrus HLB project, we aim to find out how the citurs rhizosphere microbiome change during the HLB progression. It have been observed that the HLB-infected citrus trees suffered from root decline. In previous studies, we also confirmed that the HLB infection resulted in a different microbiome pattern in citrus rhizosphere. Our goal is to track the rhizosphere microbiome alteration and further looking for the functional changes during the HLB progression. Hopefully we will know more about the association between the rhizosphere microbiome and the HLB-related root decline symptom.
In the coprophilous microbiome project, we use the elk dung as a model system to observe the succession process. During the decomposition of dung, the microbiome utlize the nutrient resources and further change the nutritional condition and temperature, which in turn drive the succession of microbiome. The elk dung pellets were collected in a series of time points. The data was been analyse through both amplicon and shotgun approaches to reveal the populational change in coprophilous microbiome.
Sicne the shotgun metagenomic analysis usually contains several steps, we also developed a snakemake workflow to process the data. Please refer to the GitHub page for more details. Note that the workflow is still under development and is subject to change.