IDRs on fungi
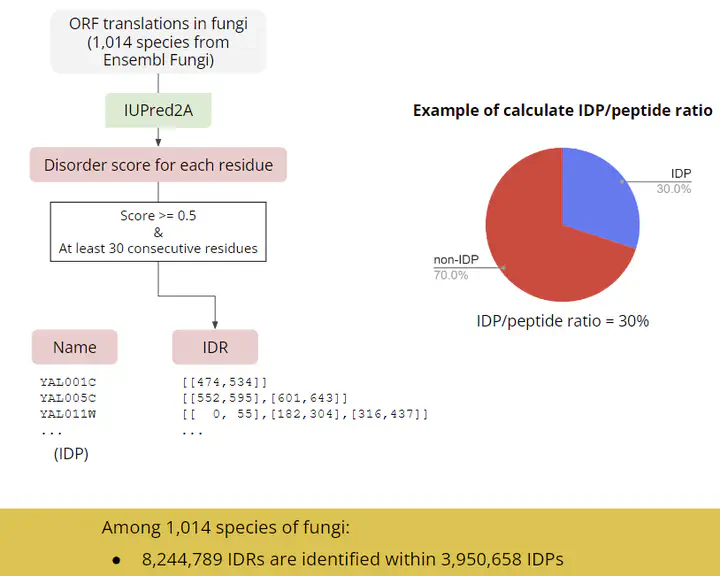
Intrinsically disodered regions (IDRs) are polypeptide segments which have flexible structure. They often lack of hydrophobic residues (which is the major driving component for protein folding) but enriched in polar/charged residues (which enhance their capability to interact with water solvent). The characteristic is used by some tools to predict the IDRs from a peptide sequence.
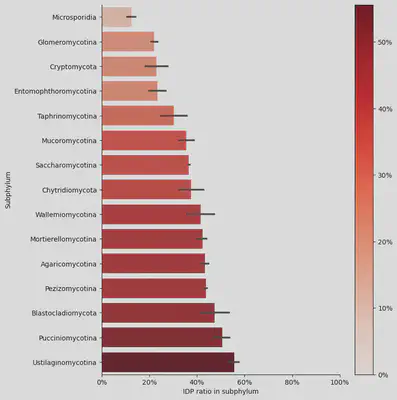
In this project, we collected the peptide sequences from 1,014 fungal species curated by Ensembl and got over 8 millions predicted IDRs by the tools IUPred2. We calculated the IDP/peptide ratio and found they are varying between subphylum - Microsporidia has only 12.5% of the peptides are IDP but more than half of the peptides in Ustilaginomycotina contains IDRs.
We further investigate the relationship between IDP/peptide ratio and genome size and generate an interactive scatterplot to visualize the result. Currently we haven’t confirm any correlation between these 2 factors.
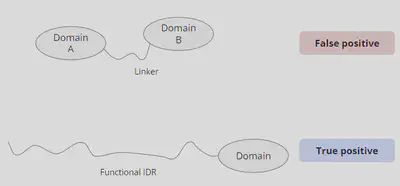
Our goal is to find out the IDRs that have specific function but major obstacle is how to effectively filter out the non-functional ones. Since all the IDR predictions are rely on IUPred2 and might contain many false positives. One interference might be linker peptides that connect each functional domain. Any linker longer than 30 bp will be classified as an IDR but probably isn’t what we want. Therefore, we need a better way to filter out all these noises and we might have chances to target those functional IDRs.