dbcanLight
Functional annotation is an important step in genome/metagenome projects. It give us hints of what these genes are capable of. One of the widely used functional database is CAZy (Carbohydrate-Active enZYmes), which describes the enzymes that degrade, modify, or create glycosidic bonds.
Currently, most of the users access the CAZy database through the tool run_dbcan, which is able to identify the CAZymes and even the potential substrates of a given peptide sequence. However, the hmmscan it use for the searching step is not very efficient and are quite time consuming when the querying/searching database is huge.
To improve the performance, I re-designed the program, utilized pyhmmer, a python bindings to HMMER3, to develop a lightweight version - dbcanLight. It reimplemented the cazyme and substrate search processes with hmmsearch and highly improve the speed.
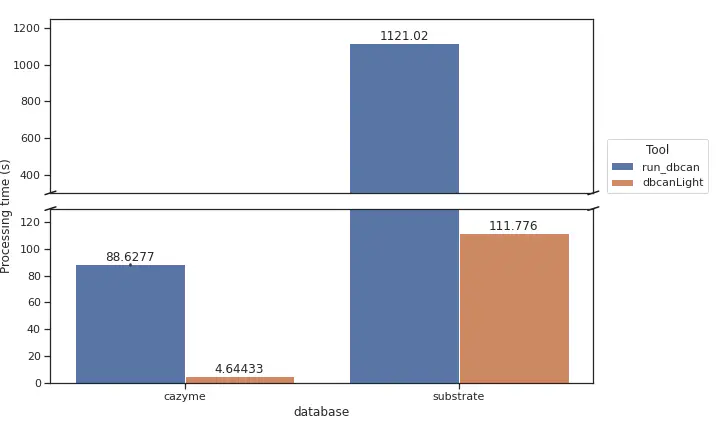
We benchmarked on a protein fasta with 9,360 sequences. 3 rounds of tests have been run on cazyme detection mode (–tools hmmer in run_dbcan and -m cazyme in dbcanLight) and 1 round of test have been run on substrate detection mode (–tools dbcansub in run_dbcan and -m sub in dbcanLight). Our tool accelates 10X in substrate detection and almost 20X in cazyme detection. Moreover, the outputs are fully compatible with the original version.
If you’re interested in dbcanLight, please refer to the GitHub page for more details.